
Decoupled Contrastive Learning for Multilingual Multimodal Medical Pre-trained Model
Paper info: Qiyuan Li, Chen Qiu, Haijiang Liu, Jinguang Gu, Dan Luo, Decoupled contrastive learning for multilingual multimodal medical pre-trained model, Neurocomputing, Volume 633, 2025, 129809, ISSN 0925-2312, https://doi.org/10.1016/j.neucom.2025.129809.
I’m excited to share our latest publication titled “Decoupled contrastive learning for multilingual multimodal medical pre-trained model” which has been published in Neurocomputing! This work represents a significant advancement in the field of medical AI, particularly in handling multilingual and multimodal data. In this blog post, I’ll walk you through the key aspects of this research and why it matters.
The Challenge: Multilingual and Multimodal Medical Data
In today’s interconnected world, healthcare systems increasingly need to serve diverse patient populations who speak different languages. Medical data itself is inherently multimodal, consisting of text reports, images (like X-rays and MRIs), and other forms of data. However, developing AI models that can effectively process this multilingual and multimodal medical data has been challenging due to:
- Data Scarcity: Quality medical data in multiple languages is difficult to obtain
- Language Imbalance: Most existing medical datasets are dominated by English and Chinese, with limited representation of other languages
- Multimodal Interaction: Effectively integrating different data types (text, images, etc.) remains technically challenging
Our Solution: 3M-CLIP Model
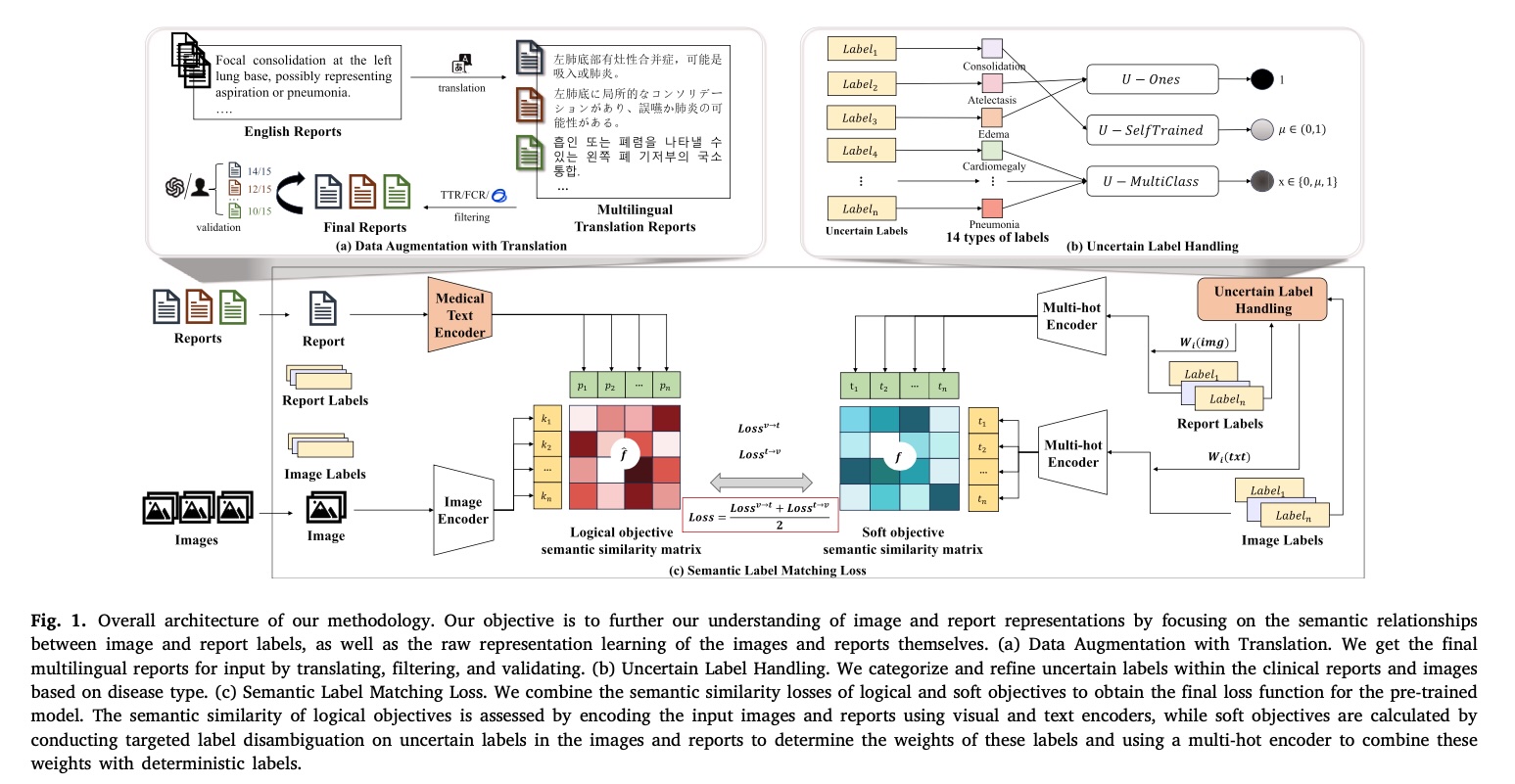
To address these challenges, we introduced the 3M-CLIP model, a novel multilingual multimodal medical pre-trained model. The key innovations of our approach include:
Data Augmentation with Translation
To address the scarcity and imbalance of multilingual medical data, we implemented a strategic data augmentation approach using machine translation. Here’s how we did it:
- Dataset Selection: We started with the MIMIC-CXR dataset, a widely-used English-language medical imaging dataset containing chest X-rays and associated radiology reports.
- Translation Process: Using the NLLB-200 model with 600M pre-training data points, we translated 216k English text data from MIMIC-CXR into 20 target languages. This gave us medical reports in 21 different languages.
- Filtering and Validation:
- We developed metrics to filter out low-quality translated data, including measuring continuous and repeated translation errors.
- We used Type Token Ratio (TTR) to assess lexical complexity and filter poor translations.
- For validation, we randomly sampled 1% of both filtered and discarded translation data and evaluated them using GLM-4-flash and human scoring across accuracy, fluency, and contextual consistency.
Model Architecture
Our 3M-CLIP model consists of three main components:
- Raw Vision-Text Encoder:
- Vision Encoder: Processes input images using a vision encoder to produce image embeddings.
- Text Encoder: Handles textual data from medical reports using an XLM-RoBERTa model fine-tuned on medical texts. This encoder captures semantic features of multilingual medical texts through masked language modeling (MLM) and translation language modeling (TLM) objectives.
- Uncertain Vision-Text Label Encoder:
- We developed targeted label disambiguation techniques to address labeling noise caused by semantic labeling errors or negative word omissions.
- We categorized and refined uncertain phrases in clinical reports based on disease types using three processing strategies (U-Ones, U-SelfTrained, U-MultiClass).
- Semantic Label Matching Loss:
- We connected images and texts based on the semantic similarity between image and text labels.
- Our loss function combines logical objectives (cosine similarity between image and text embeddings) and soft objectives (similarity between labels) to improve matching accuracy.
Targeted Label Disambiguation
We developed a method to handle labeling noise in decoupled contrastive learning. This technique:
- Identifies uncertain labels in clinical reports based on disease type
- Applies specialized processing strategies to refine these uncertain labels
- Enhances semantic similarity between different modalities (text and images)
- Improves cross-modal interactions in the model
Specific Disambiguation Strategies
We implemented three primary strategies for handling uncertain labels:
This approach maps all uncertain labels to 1, effectively treating them as positive cases. This simple strategy helps maintain data balance but may introduce some false positives.
This semi-supervised approach:
- Initially trains the model while ignoring uncertain labels
- Uses the trained model to predict labels for uncertain cases
- Incorporates these predicted probabilities as new labels for further training
This method leverages the model’s own predictions to handle uncertainty while maintaining flexibility.
This strategy treats uncertain labels as a separate category. The model outputs probabilities for three classes (0, 1, and uncertain) for each observation, allowing it to explicitly account for label uncertainty in its predictions.
Training Methodology
- Training Objectives:
- We used a dual-objective training framework combining masked language modeling (MLM) and translation language modeling (TLM) to capture semantic features of multilingual medical texts.
- Parameter Settings:
- Text encoder: Med-XLM-RoBERTa with hidden size 768, 12 attention heads, and 12 hidden layers.
- Vision encoder: Swin Transformer architecture.
- Training was conducted for 200 epochs using two A100 GPUs.
- Implementation Details:
- For images, we applied preprocessing steps including random horizontal flipping, color jittering, and random affine transformations.
- We unified the vision and text encoders to facilitate contrastive learning with an output linear projection head dimension of 514.
Loss Function specifics
Our semantic matching loss function consists of two components:
- Logical Objective Function: Calculated using the cosine similarity between image and text embeddings.
- Soft Objective Function: Based on the similarity between image and text labels.
The final loss function is the cross-entropy between these two objective functions, which helps the model learn to match images and texts more accurately across different languages and modalities.
Unified Multilingual Multimodal Framework
Our model architecture combines vision and text encoders in a unified framework that facilitates contrastive learning across different languages and data types. This allows the model to learn generalized representations of medical concepts across multiple languages and modalities.
Experimental Results
Our experiments demonstrated significant improvements over existing baseline models:
Medical Image Classification
On standard medical image classification tasks, our 3M-CLIP model showed substantial performance gains:
- CheXpert-5x200 Dataset: Accuracy improved to 0.6402 (compared to 0.5943 for the previous best model)
- MIMIC-5x200 Dataset: Accuracy reached 0.5788 (vs. 0.4856 for the previous best)
Zero-Shot Classification
In zero-shot scenarios (where the model must classify images it hasn’t specifically been trained on):
- COVID Dataset: Our model achieved 0.8428 accuracy
- RSNA Pneumonia Dataset: Accuracy was 0.8218
Fine-Tuned Classification
When fine-tuned on specific datasets, our model continued to show superior performance:
- COVID Dataset: 0.8310 accuracy
- RSNA Pneumonia Dataset: 0.8736 accuracy
Multilingual Image-Text Retrieval
In cross-lingual image-text retrieval tasks, our model outperformed previous approaches across all evaluated languages, demonstrating its strong capability in understanding medical concepts across different linguistic contexts.
Why This Matters
The 3M-CLIP model represents an important step forward in making medical AI more accessible and effective across different languages and cultural contexts. By improving the model’s ability to understand and process multilingual medical data, we can:
- Serve diverse patient populations more effectively
- Enable knowledge sharing across medical communities worldwide
- Improve diagnostic accuracy through better multimodal data integration
- Support healthcare professionals with more robust AI tools
Future Directions
While our model shows promising results, there’s always room for improvement. Future work could focus on:
- Further refining the translation quality of medical texts
- Incorporating additional modalities like audio or video data
- Developing more efficient training methods
- Exploring applications in specific medical domains
Conclusion
I’m proud of the progress we’ve made with the 3M-CLIP model in advancing multilingual multimodal medical AI. This work brings us closer to creating AI systems that can truly support healthcare providers and patients across linguistic boundaries. If you’re interested in learning more about this research, you can access the full paper here.
1 | @article{LI2025129809, |
- Title: Decoupled Contrastive Learning for Multilingual Multimodal Medical Pre-trained Model
- Author: Haijiang LIU
- Created at : 2025-03-21 16:58:36
- Updated at : 2025-10-05 11:01:24
- Link: https://github.com/alexc-l/2025/03/21/3mclip-post/
- License: © 2025 Elsevier Ltd. All rights are reserved, including those for text and data mining, AI training, and similar technologies.
